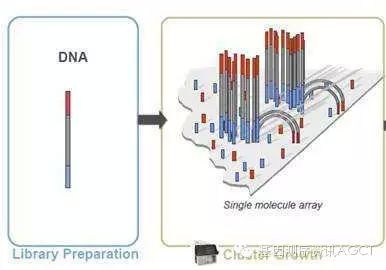
The complete second-generation sequencing workflow includes five steps: nucleic acid extraction, library preparation, cluster generation, sequencing, and data analysis. This article describes the preparation work for Illumina's NGS library before it is put into operation: nucleic acid extraction, library construction, and cluster generation. 1. Nucleic acid extraction (DNA/RNAExtraction) Extract DNA, total RNA, or small RNA from different types of samples such as cultured cells, peripheral blood, fresh frozen tissue, or paraffin embedded tissue (FFPE) using conventional methods, and then purify and quantify them. The higher the quality and completeness of DNA and RNA samples, the better. Be cautious when dealing with second-generation sequencing data obtained from degraded samples and apoptotic samples (where DNA is also damaged or degraded). Although second-generation sequencing can generate massive amounts of data, it does not consume much sample. The standard process only requires 0.1-1 μ g of DNA or RNA, while the sample size required for low input processes can be determined by each laboratory based on their own conditions and experience. There are no additional requirements for the quality of nucleic acid samples in second-generation sequencing, and overall it is the same as other molecular biology experiments. For example, the OD260/280 of DNA is between 1.8-2.0; The RIN value of RNA is>=8.0. The RIN value was determined by Agilent 2100 Bioanalyzer. In the experiment, it was found that as long as the RIN value is greater than 7, satisfactory results can be obtained. It is also recommended to use BORHEE's whole blood nucleic acid extraction reagent and matching magnetic frame here, which can improve the extraction speed and efficiency faster.
Library Construction There are various applications of second-generation sequencing, such as genome sequencing, exome sequencing, transcriptome (mRNA) sequencing, microRNA sequencing, methylation sequencing, etc ChIP-Seq、 Single cell sequencing, etc. The specific process of library preparation varies for different applications. Taking genome sequencing as an example: The construction of genomic DNA libraries is at the core of all types of second-generation sequencing library construction methods, and other processes and methods are derived from this foundation. The basic process of constructing a genomic DNA library is to first randomly break the complete long stranded DNA into short fragments with a length of about 350 bp, then connect barcode adapters at both ends of each short fragment, and then amplify the library through approximately 12 cycles of PCR. In short, the "three pronged approach" of fragmentation, ligation, and amplification involves interspersed auxiliary steps of purification and quantification. 2.1 Fragmentation By using high-pressure gas atomization, ultrasonic cavitation, ordinary ultrasonic waves, enzymes and other methods and equipment, complete long-chain DNA molecules can be broken into short fragments. The principle is that the more random the breakpoint, the better, and the more concentrated the fragment length, the better. The length of the fragment after interruption is usually around 350 bases. Of course, different applications may also require different lengths, which can be achieved by optimizing and modifying the fragmented parameter conditions. Generally speaking, the template length for SR sequencing is about 350 bases, the template length for PE sequencing is about 500 bases, and the template length for MP sequencing is about 1k-10k bases. The fragment set obtained by ultrasonic fragmentation is close to a normal distribution in length. After fragmentation, the agarose gel electrophoresis strip can be cut, or the magnetic bead purification under different buffer conditions can be combined to further narrow the length range of the fragment and accurately select the required length. Of course, it is also possible to proceed to the next step without selecting the segment length. There is no significant difference in the impact of selecting and not selecting two methods on second-generation sequencing data. The library construction pathway without selecting fragment length through gel cutting is called gel free method. 2.2 End Repair By physically breaking the obtained DNA fragments, both ends are often damaged, the structure is incomplete, and they are basically no longer flat ends, affecting the junction connection. Therefore, they need to be repaired to make them flat ends. Generally speaking, there are three types of enzymes used for end repair: enzymes with a 5 'extension, enzymes with a 5' connection, and enzymes with a 3 'extension. 2.3 3 'end with A In order to improve the connection efficiency and prevent the joint from connecting with the inserted segment in multiple directions, while reducing the interconnection between the joints, the connection between the joint and the inserted segment adopts the TA semi adhesive end connection method. This requires a protruding T base at the 3 'end of the adapter and a protruding A base at the 3' end of the insertion fragment. 2.4 Adding connectors at both ends Due to the protruding T base at the 3 'end of the adapter, when they are connected to the insertion fragment, the adapter is oriented, but the insertion fragment itself can be connected in both directions. Considering that many applications require mixing libraries from different samples for sequencing, the adapters provided by manufacturers now include barcode regions with known sequences to facilitate data splitting after sequencing is completed. These joints are numbered according to the barcode sequence, so when performing the experimental operation of joint connection, special care should be taken not to confuse the correspondence between barcode numbers and samples. In addition, different applications may have different joints in terms of sequence and structure. For example, SR sequencing and PE sequencing have different adapters and flow cells and cannot be mixed. 2.5 Purification of Connection Products Due to the efficiency issue of ligases, the ligation reaction is bound to be incomplete. In addition to the complete product with a linker at both ends, a certain proportion of various by-products will also be generated. These by-products include several situations such as one end having one end but not, both ends not having one, empty joint self connection, and free joint. The purpose of purifying the connecting products is to select complete connecting products and remove incomplete by-products. The purification method can be either cutting agarose gel electrophoresis bands or combining magnetic beads with different conditions for purification. At present, people are more inclined to use magnetic bead purification methods. 2.6 PCR Enrichment Perform 10-12 cycles of PCR on the linked product using a pair of universal primers. PCR can play two roles simultaneously. On the one hand, it can significantly increase the library size and facilitate machine sequencing; On the other hand, it can also enrich complete connection products with connectors at both ends, reducing the proportion of residual connection by-products in the library. People also have a certain degree of concern about PCR, which is that PCR may introduce data bias. If you don't like PCR, you can skip this step. The library construction pathway without PCR is called PCR free method. 2.7 Library Quality Inspection After PCR is completed, it is necessary to purify the PCR product using magnetic beads, column chromatography, or other methods, and then perform quantitative and quality control of the library to prepare for cluster generation on the machine. It is recommended to use Beckman's Ampure XP magnetic beads and BORHEE magnetic frame in combination for PCR product purification. The following methods can be selected for library quality inspection: agarose gel electrophoresis to check whether the length of PCR product meets the expectation, whether there is impurity band, whether there is pollution, whether there is primer dimer residue waiting; Agilent 2100 Bioanalyzer or other devices are used to measure the fragment length and length distribution of the library; Qubit or other devices are used to determine the mass concentration of the library. By combining Qubit and 2100 data, the molar concentration of the library can be calculated. Then dilute the library uniformly to the specified molar concentration (concentration normalization). Different sequencing platforms require different library concentrations. According to the requirements of project design, decide whether to mix different libraries and in what proportion to mix them. When mixing libraries, special attention should be paid to whether there are barcode conflicts. Never mix libraries with the same barcode together. Once mixed, they can no longer be separated and both libraries need to be reconstructed.
3.0 Cluster Generation The biochemical reaction of cluster generation occurs within a flow cell. In many fields, flow cell is translated as' flow pool '. But for second-generation sequencing, such translated words do not convey the intended meaning. Because the flow cell of second-generation sequencing is not a cup, but a row of eight long tunnels (lanes) that run through a glass sheet, highly specialized. Helpless, people have to choose not to translate and speak English directly, or use its abbreviation FC. Similarly, whether lane is translated as "channel" and whether cluster is translated as "cluster" are both awkward. Cluster generation requires specialized instruments cBot; In addition, it can also be performed on a few models of sequencers such as MiSeq and Fast HiSeq 2500. In addition to necessary preparations such as adding libraries, installing reagent plates, and setting operating parameters, cluster generation is a highly automated process that does not require manual intervention during operation. The entire process took about 4 hours. The biochemical reaction steps for cluster generation mainly include 5 steps: library template hybridization, bridge PCR, linearization, end closure, and sequencing primer hybridization. The following library processing steps involve denaturing double stranded DNA into single stranded DNA, which is unstable and needs to be freshly processed before starting cluster generation: purified (removing primer dimers, etc.), quantified, and concentration normalized DNA library (regardless of whether the original material is DNA or RNA, the library is double stranded DNA), added NaOH for alkaline denaturation, and further diluted with buffer to adjust the library concentration to the pM or nM level. A portion is divided into 8 tubes and placed on ice for machine preparation. Attention: Different sequencing platforms require different library concentrations; Some platforms have a fixed concentration on the machine and do not require adjustment; Some platforms allow for adjusting the library concentration, and we can use this method to adjust and control the data volume of each sample and lane, thereby maximizing the sequencing data volume of the entire FC. 3.1 Library Hybridization Introduce the denatured library into the flow cell using cBot or some models of sequencers, and randomly hybridize the DNA fragments of the single stranded library with the adapters distributed on the inner wall (including both upper and lower sides) of the FC channel (lane); The joint is then extended to synthesize a complementary chain, which is covalently bonded to the inner surface of the FC and fixed at a certain position on the FC. The original template single chain is washed away after NaOH alkali denaturation. 3.2 Bridge amplification The other end of the DNA single strand (complementary strand in the original library) fixed on the FC hybridizes with another junction on the inner wall of the FC due to complementary base sequences, forming a bridge like structure. Under the action of DNA polymerase, its complementary strand is synthesized. This double stranded DNA "molecular bridge" undergoes alkaline denaturation under the action of NaOH, forming two single strands, which can then hybridize with other complementary linkers to form two new "molecular bridges" and be amplified again. This process is repeated about 20 times and is called bridge PCR. Finally, each single chain template is cloned into 1000-6000 identical single chains, which are gathered together to form a cluster. Through bridge PCR, millions of clusters are randomly distributed on every square millimeter of FC inner wall, including the top and bottom layers. 3.3 Linearization The cluster formed by bridge PCR, although single stranded, contains equal amounts of sense and antisense chains, and their sequences complement each other, so there are two types of bases at each position. This will result in the presence of two colors of fluorescence in each cycle of each cluster during sequencing, making it impossible to obtain a pure fluorescence signal. Therefore, it is necessary to remove all single chains in one direction and only retain one sequence. This process is called linearization. The manufacturer pre fixes two types of connectors on the inner surface of the FC, which complement the upstream and downstream connector sequences of the library. These two types of adapters each have a site that can be cleaved by different enzymes (not restriction endonucleases), allowing for directed cleavage of single chains in one direction and removal through alkaline denaturation and buffer washing, thus completing linearization. 3.4 End closure The essence of simultaneous synthesis and sequencing is DNA strand extension. In order to ensure that only the sequencing primers are extended during sequencing, the ends of all free DNA fragments on the FC must be blocked before hybridization of the sequencing primers, so as not to interfere with the purity of the sequencing signal. Terminal closure is achieved by adding a ddNTP to the free end of a DNA fragment. 3.5 Primer hybridization Inject sequencing primers into all lanes of FC, and these primers will hybridize to the universal primer binding sites of each DNA fragment in each cluster. At this point, FC is ready and can start sequencing on the sequencer. Note: Cluster generation is best performed fresh before sequencing. The faster the cluster is completed, the better the sequencing. |  Location: Home > NEWS > Industry News
Location: Home > NEWS > Industry News Time:2024-11-30
Time:2024-11-30 Views:
Views: 
 Links:
Links: